Welcome to POSTAR3
POSTAR3 is a comprehensive database for exploring POST-trAnscriptional Regulation based on high-throughput sequencing data from 7 species, including human, mouse, zebrafish, fly, worm, Arabidopsis, and yeast. POSTAR3 is developed as the updated version of CLIPdb, POSTAR, and POSTAR2, which provides the largest binding sites collection of RNA-binding proteins and their functional annotations.
POSTAR3: an updated platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. (2021). Nucleic Acids Res.
POSTAR2: deciphering the post-transcriptional regulatory logics. (2019). Nucleic Acids Res 47, D203–D211.
POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. (2017). Nucleic Acids Res 45, D104–D114.
CLIPdb: a CLIP-seq database for protein-RNA interactions. (2015). BMC Genomics 16, 51.
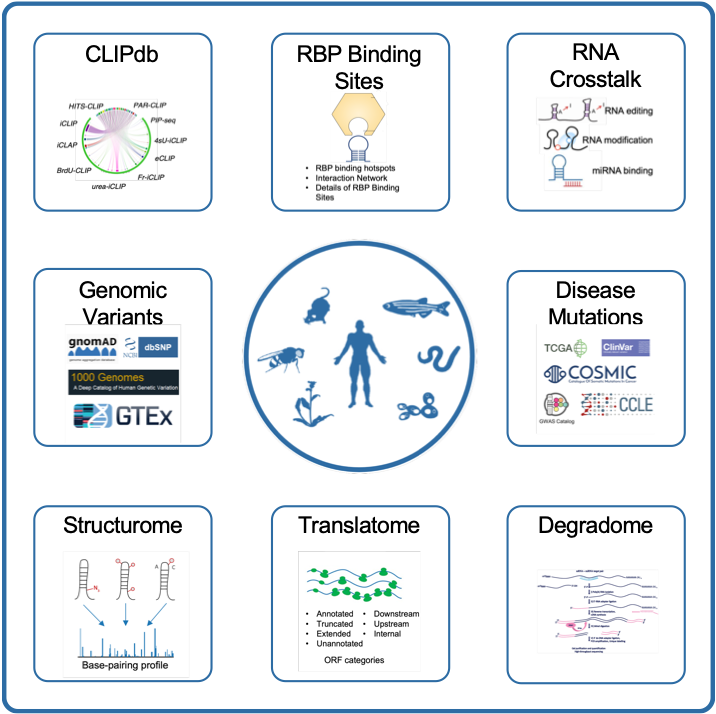
CLIPdb
CLIP-seq Database
Annotations and functions of RNA-binding proteins with CLIP-seq data.
RBP Binding
Sites

Annotations and functions of RBP binding sites.
RNA
Crosstalk
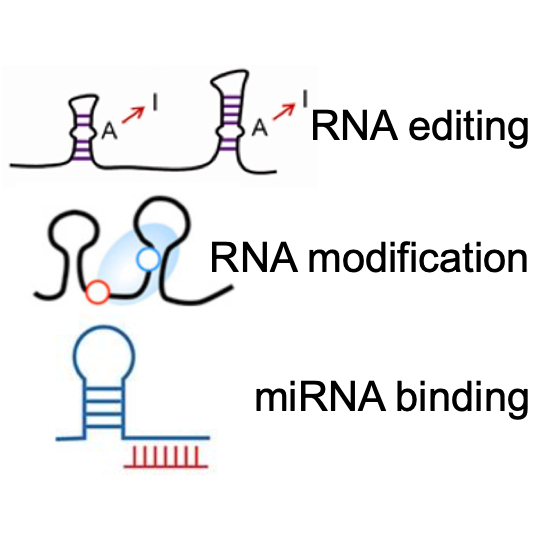
Annotations of RBP binding sites with various regulatory events.
Genomic
Variants

Annotations of RBP binding sites with genomic variants.
Disease Mutations

Annotations of RBP binding sites with disease-associated mutations.
Structurome
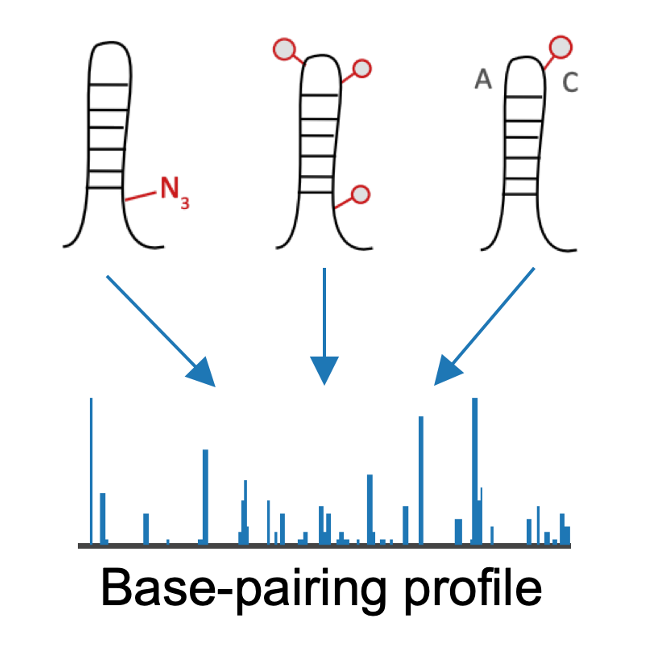
Annotations of RBP binding sites with RNA structure-seq data.
Translatome
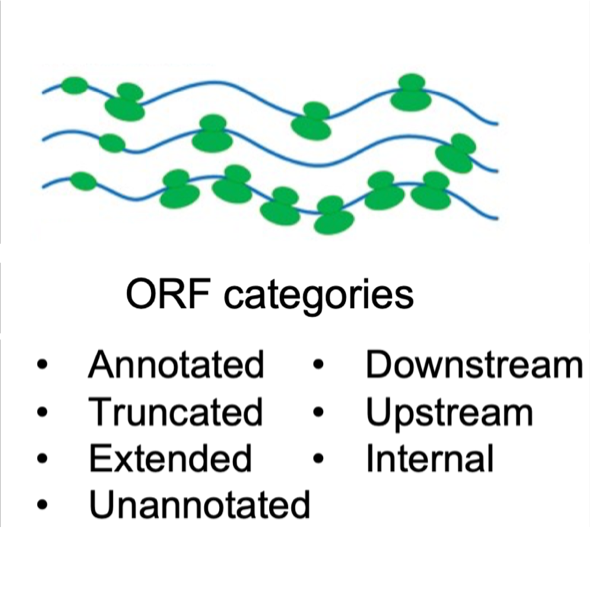
Display of translatome information with Ribo-seq data.
Degradome
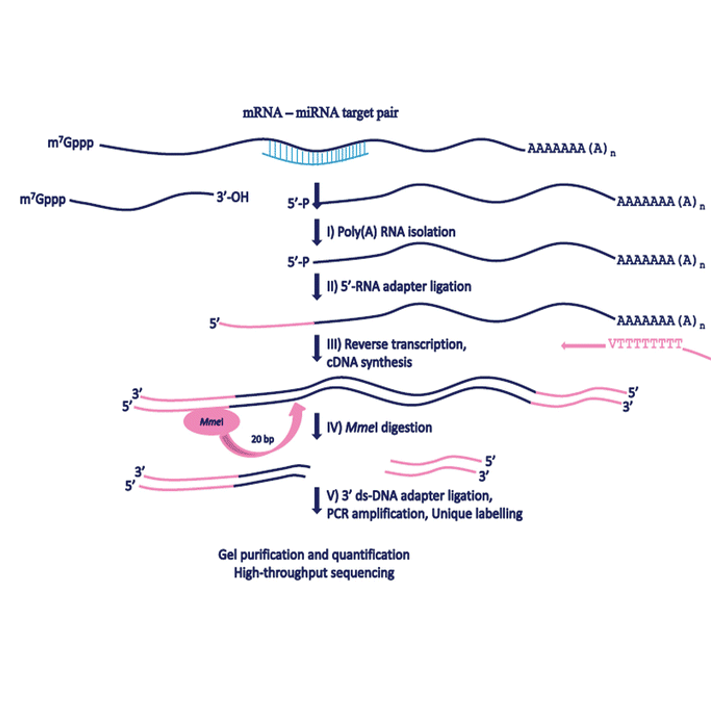
Display of degradome information with degradome-seq data.
POSTAR3 Features
POSTAR3 was released in June, 2020.
- CLIPdb: 351 RBPs with their domains, Gene Ontologies, binding sequence motifs, structural preferences, and binding sites identified from CLIP-seq experiments.
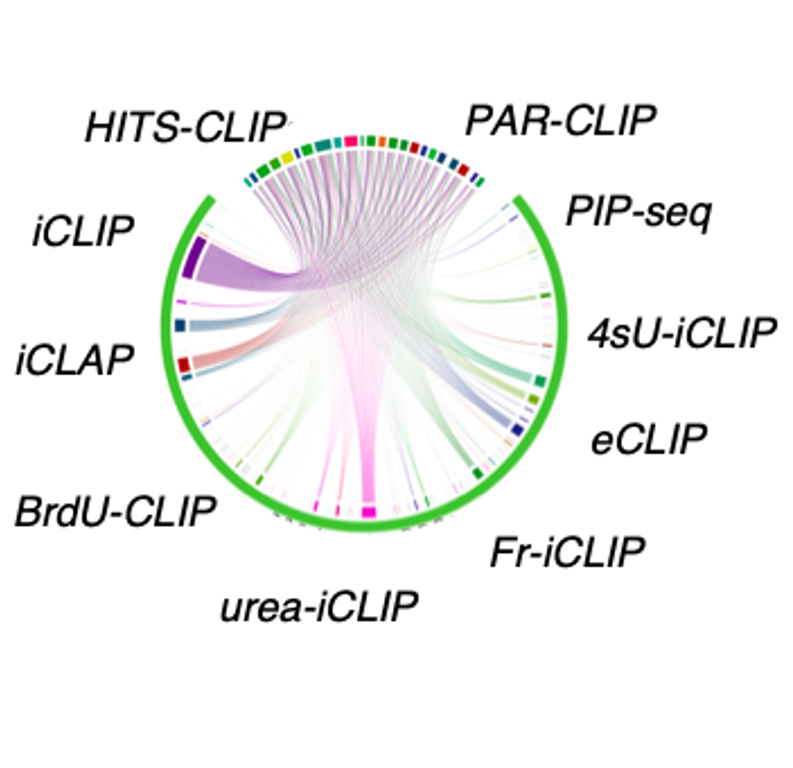
- RBP Binding Sites: RBP binding sites from 1,499 public CLIP-seq datasets covering 7 species and 10 CLIP-seq technologies.
- RNA Crosstalk: Post-transcriptional regulation annotation of RBP binding sites, including ~27 million miRNA binding sites, ~1.1 million RNA modification sites, and ~8.1 million RNA editing sites.
- Genomic Variants: Genomic variant annotation of RBP binding sites from dbSNP, 1000 genomes, Genome Aggregation Database, and eQTLs and sQTLs from GTEx.
- Disease Mutations: Disease-associated mutant annotation of RBP binding sites from ClinVar, PanTCGA, PCAWG, CCLE, GWASdb2, denovo-db, COSMIC, and HmtDB database.
- Structurome: Structurome profile of RBP binding sites from 82 public structure-seq datasets in 4 species using 6 structure-seq methods.
- Translatome: Translation dynamics of 300 public Ribo-seq datasets covering various conditions of 6 species.
- Degradome: miRNA-mediated degradation of other RNAs validated by 83 degradome-seq datasets in 4 species.
Submission Guidelines
Thank you for your contribution. All submissions will be reviewed and curated into the database as soon as possible.