RNA Crosstalk
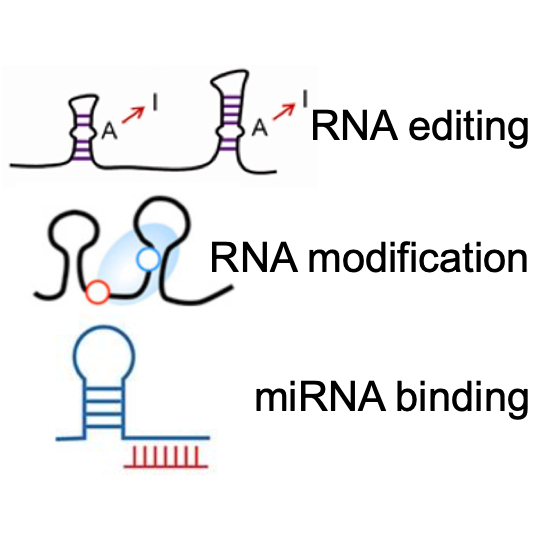
About RNA Crosstalk Module
The RNA Crosstalk module provides the interactions of RNA post-transcriptional regulations around RBP binding sites, including miRNA targets, RNA modification, and RNA editing. RBPs are involved in various kinds of post-transcription regulation events and play vital roles in these processes, so that users can investigate potential crosstalk of these regulatory events in this module.
Tips:
Search Crosstalk Module by selecting species and entering RNA name.
Species:
RNA name:
References:
1. miRanda: Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1.
2. RNAhybrid: Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451-W454.
3. psRobot: Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012;40(Web Server issue):W22-W28.
4. psRNAtarget: Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46(W1):W49-W54.
5. miRBase: Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129-138.
6. RMBase2: Xuan JJ, Sun WJ, Lin PH, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327-D334.
7. MODOMICS: Boccaletto, P., Machnicka, M.A., Purta, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303-D307.
8. N(1)-Met modification: Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature.
2016;530(7591):441-446.
9. RADAR: Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42(Database issue):D109-D113.
10. DARNED: Kiran AM, O'Mahony JJ, Sanjeev K, Baranov PV. Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 2013;41(Database issue):D258-D261.
11. Worm RNA editome: Zhao HQ, Zhang P, Gao H, et al. Profiling the RNA editomes of wild-type C. elegans and ADAR mutants. Genome Res. 2015;25(1):66-75.
12. GTEx RNA editome (human, mouse): Tan MH, Li Q, Shanmugam R, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550(7675):249-254.
13. Zebrafish RNA editome: Ilana Buchumenski, Karoline Holler, Lior Appelbaum, et al. Systematic identification of A-to-I RNA editing in zebrafish development and adult organs. Nucleic Acids Res.
2021;49(8):4325-4337.