Degradome
About Degradome Module
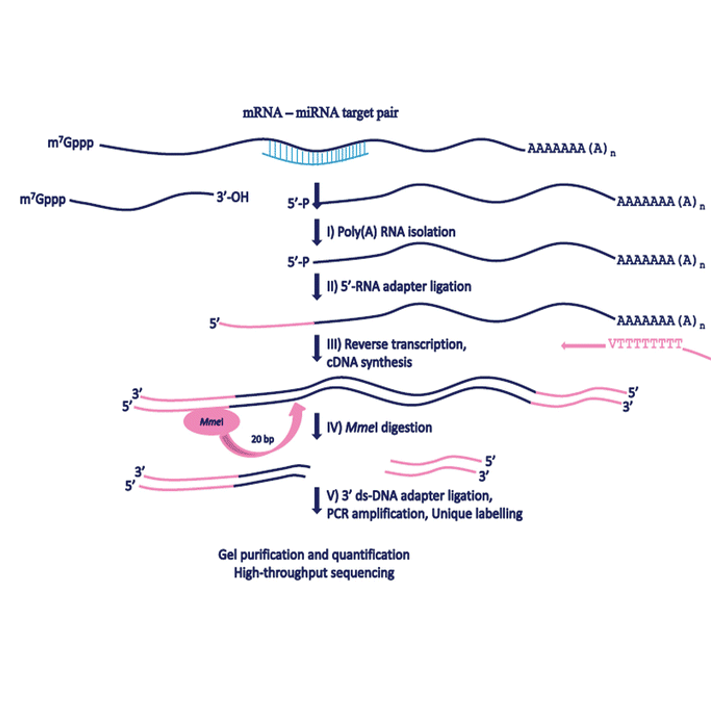
The degradome module provides the binding between small RNA and other types of RNA which leads to degradation of the other target RNA validated by degradome-seq data. A miRNA usually can exert post-transcriptional control over a broad set of cellular RNAs. User can get detailed information about every validated sRNA-fragment pair.
Tips:
Search Degradome Module by selecting species, RNA type and entering RNA name.
Species:
Search RNA type:
Search RNA name:
References:
1. PARE: German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc.
2009;4(3):356-362.
2. Degradome: Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18(10):758-762.
3. GMUCT: Gregory BD, O'Malley RC, Lister R, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854-866.
4. PAREsnip2: Thody J, Folkes L, Medina-Calzada Z, Xu P, Dalmay T, Moulton V. PAREsnip2: a tool for high-throughput prediction of small RNA targets from degradome sequencing data using configurable targeting rules. Nucleic Acids Res. 2018;46(17):8730-8739.
5. Gene Ontologies: Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-29.
6. miRBase: Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res.
2006;34(Database issue):D140-D144.