About
| Species | Human | Mouse | Fly | Worm | Arabidopsis | Yeast | Zebrafish |
| Reference genome version | hg38 | mm10 | dmel-r6.18 | ws235 | TAIR10 | R64-1-1 | danRer11 |
| Software | Description | Version | Reference |
| CLIP-seq read mapping | |||
| Bowtie | A fast alignment tool used for CLIP-seq read mapping | v1.0.0 | PMID 19261174 |
| Novoalign | A program used for HITS-CLIP and iCLIP read mapping | v3.00.05 | Link |
| CLIP-seq peak calling | |||
| Piranha | Peak calling algorithm for all types of CLIP-seq dataset | v1.2.1 | PMID 23024010 |
| PARalyzer | Peak calling algorithm for PAR-CLIP dataset only | v1.1 | PMID 21851591 |
| CIMS | Peak calling algorithm for HITS-CLIP dataset only | v1.0.4 | PMID 24407355 |
| CITS | Peak calling algorithm for iCLIP dataset only | v1.0.4 | PMID 24613350 |
| CTK | Peak calling algorithm for HITS-CLIP and BrdU-CLIP dataset only | v1.1.3 | PMID 27797762 |
| Clipper | Peak calling algorithm for HITS-CLIP only | v0.1.4 | PMID 24213538 |
| MiClip | Peak calling algorithm for PAR-CLIP only | v1.3 | PMID 24714572 |
| PureCLIP | Peak calling algorithm for iCLIP, eCLIP and related dataset only | v1.3.1 | PMID 29284540 |
| Predicting sequence motifs of RBP binding sites | |||
| MEME | Identify sequence motifs of RBP binding sites | v4.9.1 | PMID 7584402 |
| HOMER | Identify sequence motifs of RBP binding sites | v4.10 | PMID 20513432 |
| WebLogo | Visualize sequence motifs | v2.8 | PMID 15173120 |
| Predicting structural preferences of RBP binding sites | |||
| RNApromo | Identify enriched structural elements of RBP binding sites | v3 | PMID 18815376 |
| RNAcontext | Identify structural preferences of RBP binding sites | Jan 2016 | PMID 20617199 |
| Predicting miRNA-binding sites | |||
| miRanda | Predict miRNA targets by evaluating sequence complementarity and thermodynamic stability of RNA local alignment | v3.3a | PMID 14709173 |
| RNAhybrid | Predict miRNA targets by finding the minimum free energy hybridization of a long and a short RNA | v2.1.1 | PMID 16845047 |
| psRobot | Predict miRNA targets by identifying smRNAs with stem-loop shaped precursors among batch input data and predicts their targets in plants | v1.2 | PMID 22693224 |
| psRNAtarget | Predict miRNA targets by integrating a predefined scoring schema and evaluating target site accessibility in plants | v2 | PMID 29718424 |
| Transcriptome analysis | |||
| TopHat | A fast splice junction mapper for RNA-seq reads | v2.0.10 | PMID 19289445 |
| Cufflinks | Assemble transcripts and estimates their abundances for RNA-seq data | v2.1.1 | PMID 20436464 |
| Translatome analysis | |||
| RiboWave | A wavelet-based algorithm for identifying actively translated ORFs | v1.0 | PMID 29945224 |
| RiboCode | An algorithm for identifying ORFs from Ribo-seq data | v1.2.11 | PMID 29538776 |
| RiboTaper | A multitaper spectral-based approach for detecting actively translated ORFs | v1.3 | PMID 26657557 |
| ORFscore | A metric for exploring high-resolution footprinting with frame preference to identify actively translated ORFs | NA | PMID 24705786 |
| RibORF | A support vector machine-based classifier for identifying actively translated ORFs. | v0.1 | PMID 26687005 |
| Structurome analysis | |||
| RNAstructure (Fold) | An algorithm for folding RNA sequence into secondary structure with structure-seq restraints | v6.2 | PMID 20230624 |
| ViennaRNA (RNAfold) | An algorithm for folding RNA sequence into secondary structure with structure-seq restraints | v2.4.9 | PMID 22115189 |
| Degradome analysis | |||
| PAREsnip2 | An algorithm for predicting and evaluating degradome target | v4.5 | PMID 30007348 |
| Other tools | |||
| FASTX-Toolkit | A collection of tools for CLIP-seq data preprocessing | v0.0.13.2 | PMID 21278185 |
| fastp | Used for Ribo-seq data preprocessing | v0.20.0 | PMID 30423086 |
| Cutadapt | Used for degradome-seq data preprocessing | v2.8 | Link |
| BEDTools | Work for the comparison, manipulation and annotation of genomic features in BED and GTF format | v2.17.0 | PMID 20110278 |
| Resource | Description | Version | Reference |
| CLIPdb | |||
| ENCODE eCLIP | Provide RBP binding sites identified using eCLIP technology | Apr. 2020 | PMID 32252787 |
| RBP annotation | Information about RBP gene symbol and RNA-binding domains | NA | PMID 25365966 |
| Gene Ontologies | Define concepts/classes used to describe gene function, and relationships between these concepts | Sep. 2020 | PMID 10802651 |
| Pfam | Sequence motif of RBP | v33.1 | PMID 30357350 |
| RBP Binding site & Translatome | |||
| GENCODE | Human and mouse genome annotation | human v27; mouse v7 | PMID 22955987 |
| Flybase | Fly gene annotation | dmel-r6.18 | PMID 18641940 |
| WormBase | Worm gene annotation | ws235 | PMID 19910365 |
| TAIR | Arabidopsis gene annotation | TAIR10 | PMID 22140109 |
| SGD | Yeast gene annotation | R64-1-1 | PMID 12519985 |
| ZFIN | Zebrafish gene annotation | GRCz11 | PMID 33170210 |
| PhastCons | Conservation scores for alignments of multiple genomes | UCSC database | PMID 16024819 |
| PhastCons (Arabidopsis) | Conservation scores for Arabidopsis | NA | PMID 23150631 |
| phyloP | Basewise conservation scores of multiple genomes | UCSC database | PMID 19858363 |
| Gene Ontologies | Define concepts/classes used to describe gene function, and relationships between these concepts | Sep. 2020 | PMID 10802651 |
| circBase | circRNA annotation, including genomic location, expression value, and other information | NA | PMID 25234927 |
| RNA Crosstalk | |||
| miRBase | Published miRNA sequences and annotation | v21 | PMID 16957372 |
| RMBase2 | RNA modification sites identified from high-throughput sequencing datasets | v2.0 | PMID 29040692 |
| RADAR | A-to-I editing sites collected from published datasets and identified from high-throughput sequencing datasets | v2 | PMID 24163250 |
| DARNED | RNA editing sites collected from published datasets | NA | PMID 23074185 |
| RNA editome (worm) | RNA editomes of wild-type C.elegans | NA | PMID 25373143 |
| Genomic Variation | |||
| dbSNP | Public archive for genetic variation | NA | PMID 33095870 |
| 1000 genomes | Public archive for genetic variation of 1000 human around the world | NA | PMID 26432245 |
| gnomAD | Genome Aggregation Database | NA | PMID 32461654 |
| GTEx eQTL | Genotype-Tissue Expression program, expression Quantitative Trait Loci | NA | PMID 29022597 |
| GTEx sQTL | Genotype-Tissue Expression program, splicing Quantitative Trait Loci | NA | PMID 32913098 |
| Disease Mutations | |||
| GWASdb | Comprehensive data curation and knowledge integration for GWASs | v2 | PMID 26615194 |
| ClinVar | Relationships between human variations and phenotypes, with supporting evidence | Apr. 2020 | PMID 31777943 |
| COSMIC | Comprehensive resource for information on somatic mutations in human cancer | v76 | PMID 30371878 |
| TCGA whole-genome SNVs | Whole-genome somatic mutations in human cancer (TCGA) | NA | PMID 23945592 |
| TCGA whole-exome SNVs | Whole-exome somatic mutations in human cancer (TCGA) | NA | PMID 29596782 |
| CCLE | Cancer Cell Line Encyclopedia | NA | PMID 31068700 |
| Denovo-db | A compendium of human de novo variants | v1.6.1 | PMID 27907889 |
| HmtDB | a Human Mitochondrial Genomic Resource Based on Variability Studies | NA | PMID 27899581 |
| Degradome | |||
| miRBase | miRNA annotation in degradome | NA | PMID 25234927 |
| Gene Ontologies | Define concepts/classes used to describe gene function, and relationships between these concepts | Sep. 2020 | PMID 10802651 |
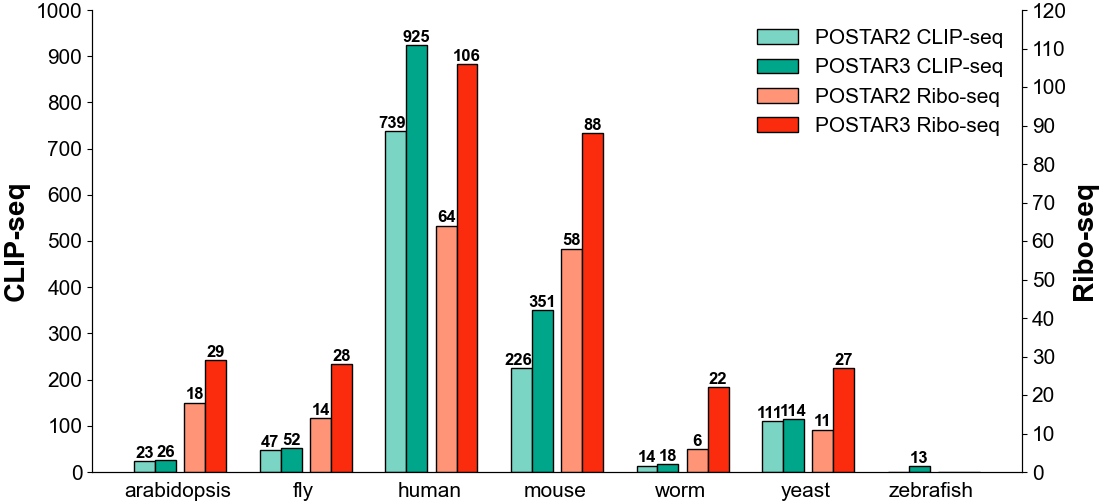
Number of CLIP-seq and Ribo-seq datasets in 7 species, compared with POSTAR2
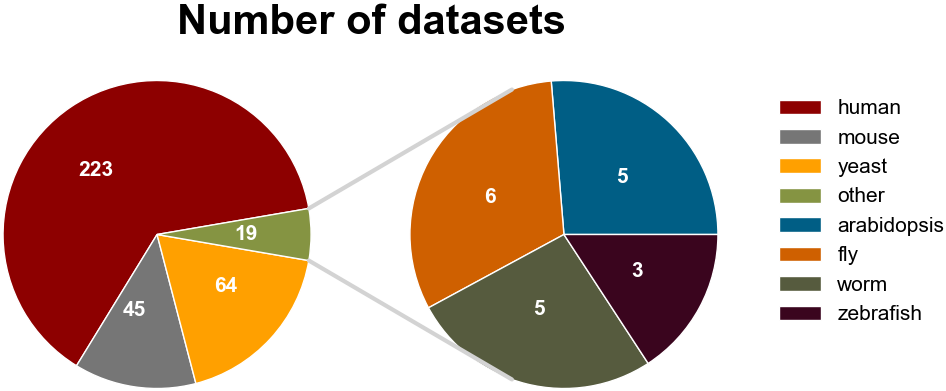
Number of CLIP-seq datasets using different technologies
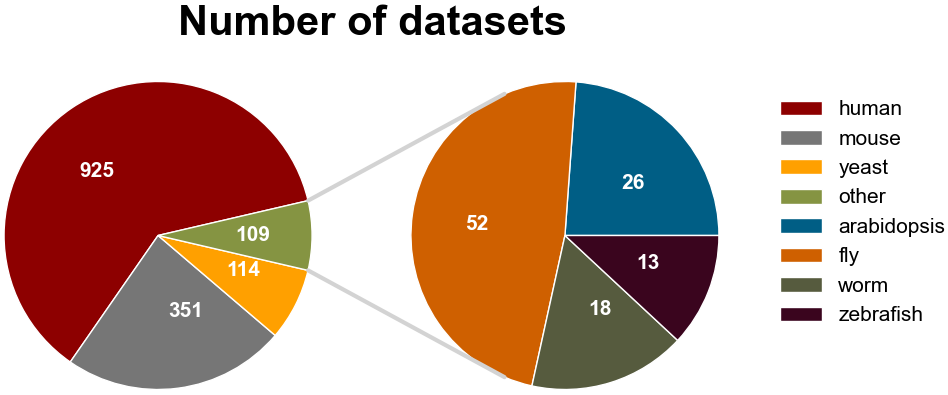
Number of RBPs in 7 species
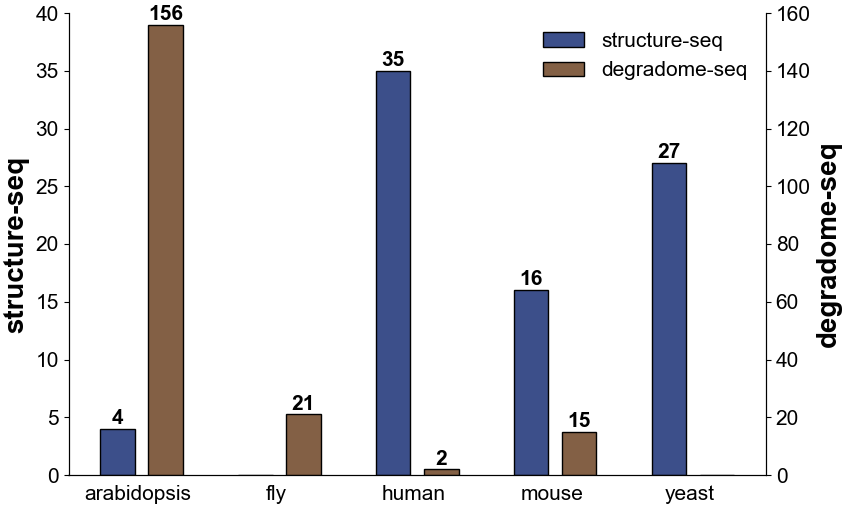
Number of structure-seq and degradome-seq datasets curated in POSTAR3
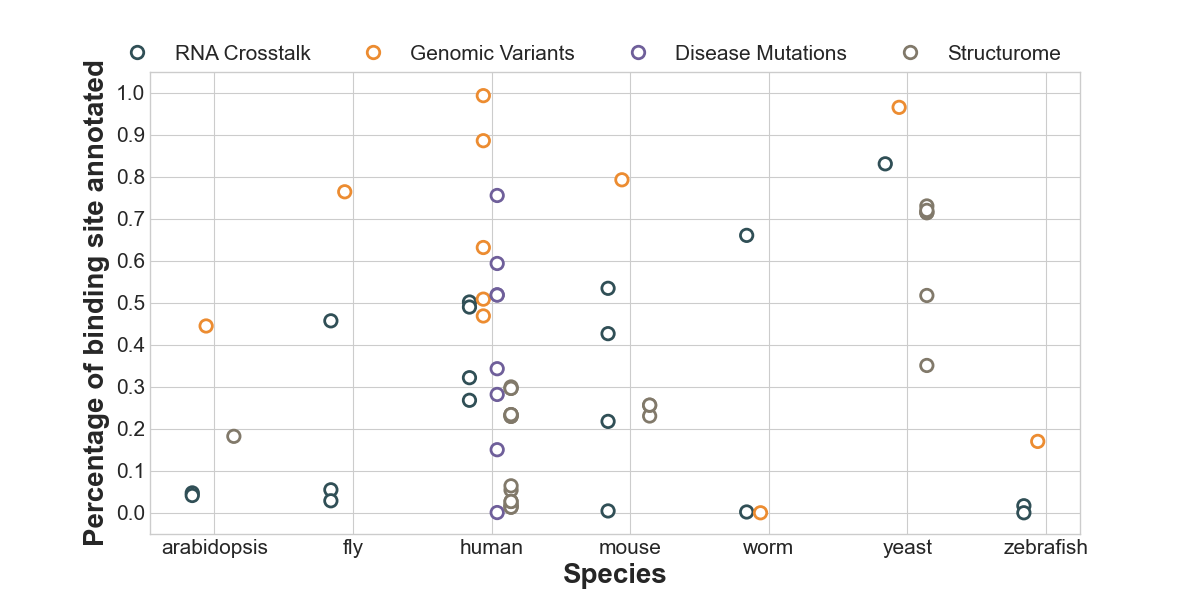
Annotation status of RBP binding sites in different modules
| Chrome | Firefox | Safari | Microsoft Edge | Opera | Android Browser & WebView | Internet Explorer | |
| Android | Supported | Supported | N/A | Supported | Supported | Android v5.0+ supported | N/A |
| iOS | Supported | Supported | Supported | Supported | Supported | N/A | N/A |
| Windows 10 Mobile | N/A | N/A | N/A | Supported | Supported | N/A | N/A |
| Mac | Supported | Supported | Supported | Supported | Supported | N/A | N/A |
| Windows | Supported | Supported | N/A | Supported | Supported | N/A | Supported, IE10+ |